pyOpenBCI library ?
Hello everyone, I have recently been using cEEGrid and Cyton+Daisy to do some small experiments, and transmit the collected signal to python for further recognition processing. However, sometimes when the electrodes are not connected to the ear or are not connected to the board at all, I can see the signal change in python( but this does not happen every time.) Pls is there something wrong with my board?Or is there a problem with my signal being transmitted to python?

Comments
More precisely, the experimental process utilizes the pyOpenBCI library for data output
Hello 'start',
How does the signal (Time Series) look with the OpenBCI GUI ALONE? If it looks good there, but looks strange in Python, then the issue is somewhere other than hardware.
Another point is that https://github.com/openbci-archive/pyOpenBCI is no longer supported and has not been maintained for years (5 years!)
You need to be using the Brainflow library instead:
https://brainflow.org/
https://brainflow.readthedocs.io/en/stable/
https://brainflow.readthedocs.io/en/stable/Examples.html
Regards, William
It's so kind of you to notice and answer my questioin.
According to your suggestion, firstly, I tried to observe the signal with GUI alone. It looked normal(with no wave) and finally stopped due to the lack of fluctuation. However, when using Brainflow library to collect the signals(board.get_board_data()), I can see different fluctuations in differrent channels, among which the highest fluctuation can be up to ±100μV.
Does it mean that there is nothing wrong with the hardware? If so, is there anything left by me during the process of signal collecting?
Please forgive me for my terrible English. Thanks for your kindness. Best wishes!
Remember that the GUI display window / subwindows, have already pre-filtered the data being shown. You must apply similar filters to the data you collect with Brainflow, as that data stream is RAW, no filtering. Just as the GUI recordings are also raw.
A typical set of filters to apply might be a bandpass filter from say .5 Hz to say 45 Hz. Then also a notch filter at your mains frequency, to eliminate that strong source of artifact generation.
Thanks for your answer. Acutally, after using the filter, the signal looks much more normal when testing without electrodes.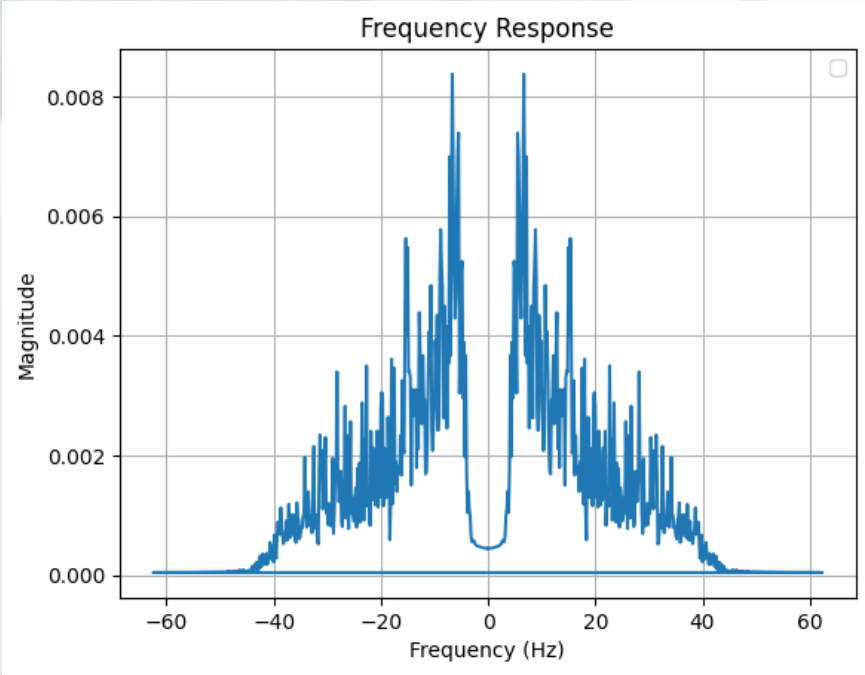

However, I seem to have another problem--I CANNOT SEE SSVEP. It seems that there are many noises within the signals .
To clarify more clearly, firstly, please allow me to briefly introduce the current situation.
The instrument I choose for my test is cyton+daisy and cEEGrids(the availability can be verified by this article https://ieeexplore.ieee.org/document/8758838). And I try to transform SSVEP with this set of instrument.
For stimulus images, we wrote it using Python's pyqt and displayed it on a computer, but this was limited by both the screen refresh rate (165Hz in ours) and the timer accuracy (1ms), and we guessed that this caused the flicker frequency to be unstable and the target peak on the FFT was submerged, and we wanted to know how likely this guess is? and we wanted to know if there was a better way to write stimulus images to get a stable frequency of stimulus.
Besides, I went through the previous sessions and found that my situation may be similar to https://openbci.com/forum/index.php?p=/discussion/3450/failing-to-see-any-ssvep-is-my-raw-data-bad-quality
I would like to ask, does the cyton board have the same REF and differential mode switching as Ganglion?
Thank you for your patience. As I really want to perform the test successfully and try something cool, your answer will be really helpful for me .
(The following is a signal collected from the 13.75Hz flash stimulation, I hope to give you some reference)!
The following figure shows our impedance test, the red channel is mainly due to the sideburns, and most of the channel impedance is quite ideal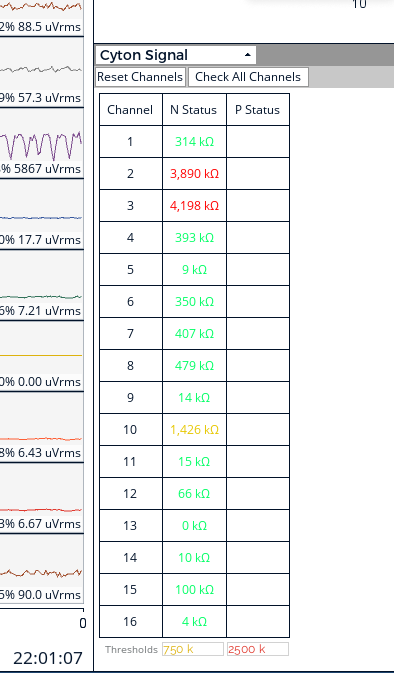
re "when testing without electrodes"
Electrodes on scalp are always needed for valid EEG. Without that, much environmental / EMF noise can be received, depending on your particular situation.
re: SSVEP stimulation frequency accuracy
Yes you are correct. programmatic generation may not be optimal, as processor scheduling is CPU and GPU load dependent. As you show, other users have encountered the same difficulty. You may find some measurement of your accuracy using the MindAffect diagnostic:
https://mindaffect-bci.readthedocs.io/en/latest/stimulus_time-lock_test_tutorial.html
re: cVEP or SSVEP electrode locations
Optimal location is most likely occipital lobe. MindAffect just used 4 channels.
https://mindaffect-bci.readthedocs.io/en/latest/printing_guide.html
re: Cyton vs Ganglion
Both boards can be configured with a common reference. Ganglion board switches default to 'differential' mode, not common reference.
https://mindaffect-bci.readthedocs.io/en/latest/installation.html#framerate-check
Using mindaffect check, the frame rate is fine, I tried to check stimulus time-lock quality, but the result is can not connect to utopia server (this may be related to the network in my region). I switched to using pyglet.clock.schedule_interval as a timer, do you know if this is possible, and in previous experience, can it meet the accuracy requirements?
Frame rate check is ONLY checking frame rate, with no photodiode (stimulus) input comparison. The other page I linked to uses the photo sensor.
https://mindaffect-bci.readthedocs.io/en/latest/stimulus_time-lock_test_tutorial.html
In general, I believe the cVEP (code based VEP) is much superior and faster than traditional old-style SSVEP. Have you watched the demos?